Bioengineer
3d
370
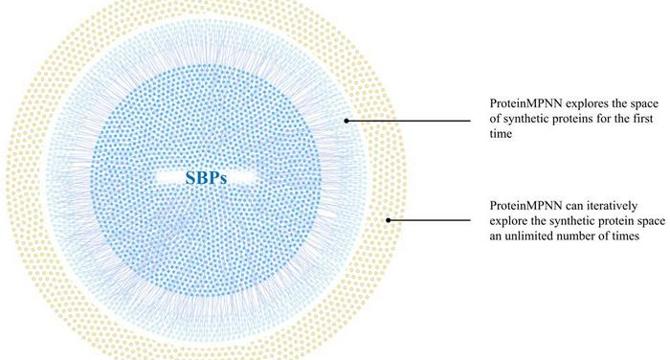
Image Credit: Bioengineer
Enhancing the Diversity of Synthetic Binding Proteins Through a Deep Learning Framework: Introducing ProteinMPNN
- Protein engineering faces challenges due to limitations of traditional methods like site-directed mutagenesis and directed evolution, hindering the exploration of therapeutic options beyond existing proteins.
- The advent of deep learning-based frameworks, such as ProteinMPNN, has revolutionized protein design by expanding sequence space for synthetic binding proteins (SBPs).
- ProteinMPNN utilizes machine learning for stability and folding predictions, offering potential advancements over energy function-based approaches.
- Research led by Dr. Weiwei Xue successfully utilized ProteinMPNN, resulting in SBPs with enhanced properties like solubility and stability, outperforming conventional techniques.
- Bioinformatics analysis revealed that ProteinMPNN-derived sequences showed improved properties compared to original SBPs, showcasing the framework's effectiveness.
- The study identified eight scaffolds with enhanced solubility and stability, crucial for synthetic binding protein functionality, offering opportunities for addressing clinical challenges like targeted drug delivery.
- The integration of deep learning into protein design through ProteinMPNN could lead to personalized therapies by uncovering patterns inaccessible to traditional methods.
- The research signifies a unique interdisciplinary collaboration between deep learning and molecular biology, advancing solutions for complex biological challenges.
- ProteinMPNN's potential impact extends to developing personalized treatments for diseases like cancer and autoimmune disorders, overcoming resistance to conventional therapies.
- Future studies will focus on refining predictive models and expanding datasets to improve accuracy and applicability, ushering in advancements in protein design and therapeutic development.
Read Full Article
22 Likes
For uninterrupted reading, download the app