Bioengineer
1w
156
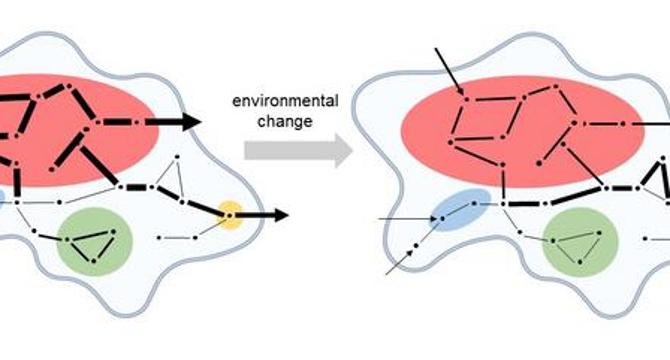
Image Credit: Bioengineer
Kinetic Coupling: A Major Breakthrough in Decoding Biochemical Networks
- A collaborative effort between researchers at the University of Potsdam and the Max Planck Institute introduces kinetic modules in biochemical networks, reshaping our understanding of cellular systems' function and stability.
- Published in Science Advances, the research bridges the gap between network structure and dynamic behavior to explain how biochemical systems maintain metabolite concentrations amidst environmental fluctuations.
- Kinetic modules are defined by their interdependence in reaction rates, showcasing how cellular robustness depends on the synchronization of biochemical reactions.
- The study, led by Zoran Nikoloski, emphasizes the clinical implications of understanding kinetic modules, offering new avenues for therapeutic intervention and metabolic engineering.
- By analyzing diverse metabolic networks, the researchers reveal how kinetic modules stabilize metabolite concentrations, providing insights into cellular adaptability and stability.
- Kinetic modules act as intrinsic sources of concentration robustness, buffering metabolic outputs from perturbations caused by environmental changes or genetic variations.
- The introduction of kinetically defined modules paves the way for advancements in systems biology and synthetic biology, aiding in optimizing metabolic pathways and developing novel therapeutic strategies.
- These findings also shed light on evolutionary perspectives, suggesting that kinetic modules are conserved functional units shaped by both genetic selection and dynamic requirements for stability.
- Illustrations accompanying the research visually represent biochemical networks with colored kinetic modules and varying arrow thickness to showcase dynamic relationships within metabolic interactions.
- Understanding and manipulating kinetic modules could revolutionize precision medicine by targeting metabolic dysregulation observed in diseases like cancer, diabetes, and metabolic syndromes.
Read Full Article
9 Likes
For uninterrupted reading, download the app