Bioengineer
2d
141
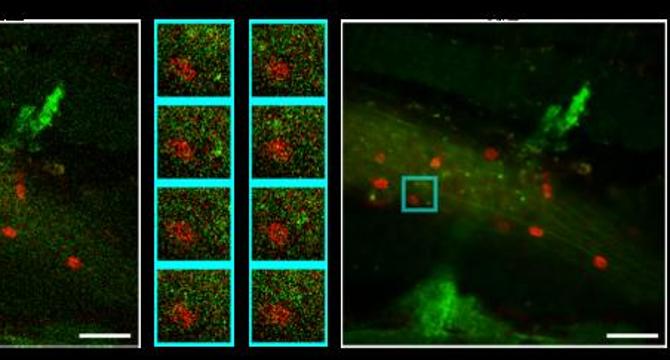
Image Credit: Bioengineer
Revolutionizing High-Speed Dynamic Fluorescence Imaging with Deep Learning-Enhanced Denoising Techniques
- Researchers introduced a groundbreaking method in fluorescence microscopy to address image degradation from noise in dynamic in vivo imaging.
- The self-supervised deep learning approach Temporal-gradient empowered Denoising (TeD) was published in PhotoniX.
- TeD aims to improve capturing and analyzing high-speed biological processes obscured by noise effectively.
- The model incorporates a temporal gradient-based attention mechanism to enhance fluorescence image quality without requiring pristine reference images.
- TeD selectively utilizes relevant spatiotemporal features to preserve moving structures like blood cells, showcasing promise in various imaging modalities.
- Validation confirmed TeD's ability to recover fine structural details under dynamic conditions, enhancing signal-to-noise ratio and structural fidelity compared to traditional methods.
- The method enables better quality fluorescence images, aiding in deeper exploration of biological processes' spatiotemporal dynamics.
- TeD's flexibility in real-world scenarios without clean reference images opens avenues for progress in biological imaging, impacting developmental biology and neuroscience.
- Researchers highlight TeD's importance in advancing scientific understanding of dynamic biological processes and its potential applications in various research areas.
- The study emphasizes the significance of machine learning in scientific research, setting the stage for advancements in biological imaging and beyond.
Read Full Article
8 Likes
For uninterrupted reading, download the app